Cell and Molecular Biology Application. The Wasabi project
1. About Bioinformatics & Cellular Molecular biology
1.1 Bioinformatics
Bioinformatics is a research where physicists, mathematicians, biologists and computer scientists work together on a the problem of biology.
The bioinformatics recherches focus on genome analysis, modeling of animal population evolution in a specific environment. This analysis involves molecular modeling, image analysis, genome assembly and phylogenetic tree reconstitution.
On the bioinformatics environment we speak about « in-silico biology » by analogy with in vitro or in vivo.
1.2 Cellular and Molecular biology
Cellular and Molecular biology is a scientific discipline that brings together genetics, biochemistry and physics. Its purpose is the understanding of the functioning mechanisms of the cell at the cellular or molecular level.
2. What is Wasabi Project ?
| partners / contributors | ||
|---|---|---|
| Arnaud Bonnafoux | LBMC, Laboratoire de Biologie et Modélisation de la Cellule | |
| Olivier Gandrillon | LBMC, Laboratoire de Biologie et Modélisation de la Cellule | |
2.1 Wasabi
The WASABI (Wave Analysis Based Inference) Project is an application of bioinformatics developed by biologist Arnaud Bonnafoux from the laboratory LBMC of the ENS de Lyon. This application was designed with the objective of identifying the gene network (GRN) that controls the behavior of a cell.
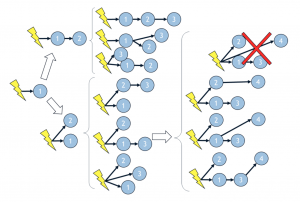
Figure 1: Scheme of identification of the candidate networks
The Wasabi project is set up to identify in silico networks of candidate genes that behave similarly to in vitro networks. The goal of the Wasabi project is to understand the mechanisms of cellular behavior but also to have the knowledge to predict the reactions of cells according to the situations encountered.
Simulations involve acting on candidate gene networks. Following each simulation, the identification of candidate networks is done by integrating the new genes into the new candidate networks generated (Figure 1).
At each stage of identification, networks are calibrated and then selected or rejected according to a set of criteria and mathematical formulas to compare their similarities to in-vitro data.
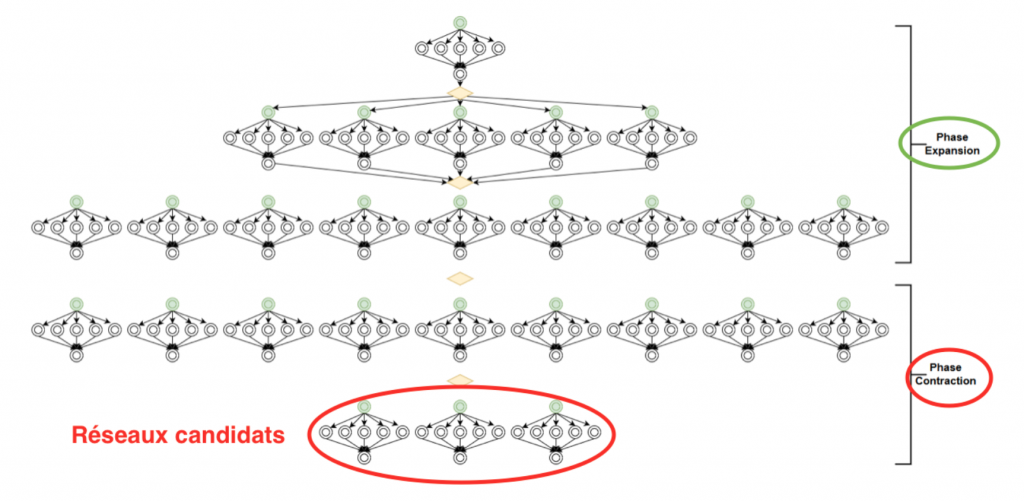
Figure 2: Diagram of the dynamic workflow generated by the iterative identification process
The workflow required a lot of memory if we launch it on the same machine. Setting up a dynamic workflow on several machines using a client-server is therefore necessary.
2.2 The DIET Client-Server
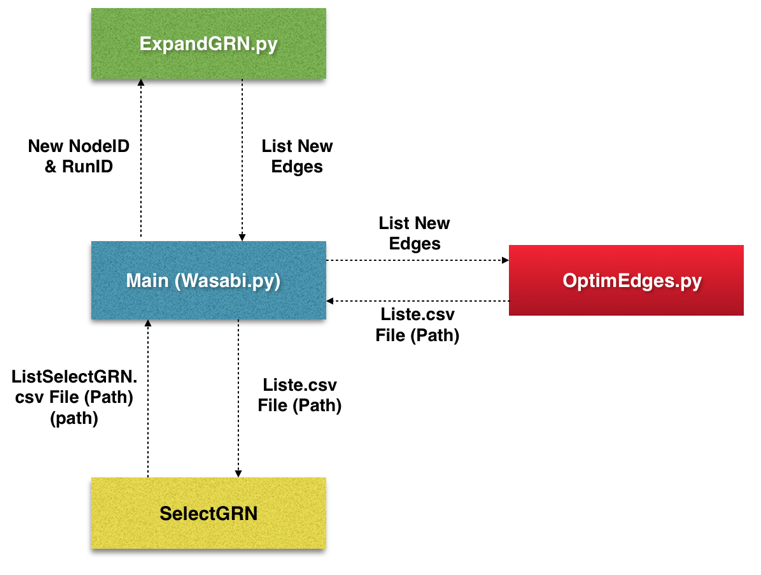
Figure 3: The final restructuring of the Wasabi project
Realization of the restructuring of the Wasabi application to interface with DIET (client-server oriented). It was necessary to review the code architecture in order to make it usable and optimal on the distributed computing middleware.
It was therefore necessary to consider splitting the code into three distinct parts (Figure 3) in order to transform the main (Wasabi.py) into a client. We distinguish in green a part corresponding to the development of the network of genes. The red part is used to optimize the calculations (comparison with in vitro data). And the yellow part is the selection of the best gene networks.
2.3 Interface
The interface is the link between the client-server and Wasabi application and allows to hide the entire DIET part to users.
The WebBoard has two main parts: one corresponding to the launch of the Wasabi simulations and another allowing access to a table of jobs and the results obtained following the simulations.
The simulation part consists of retrieving the data transmitted by the user via the form and transmitting them to the client-server in order to execute the Wasabi application (Figure 4).
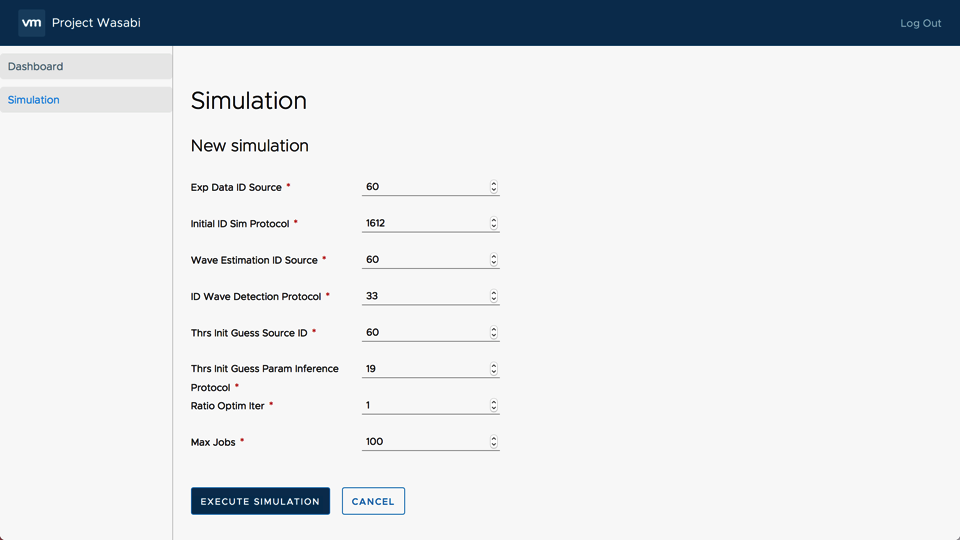
Figure 4: Screenshot of the Webboard form
Once the simulation is launched, a new job is created and appears in the job board. For each job, we find the date and time of the beginning and the end of the simulation. The table also shows the progress of the simulation and its status (initialized, in progress, completed, …) (Figure 5).
To access all the results of a job, select it. Then all the data corresponding to the selected job are displayed (Figure 5).
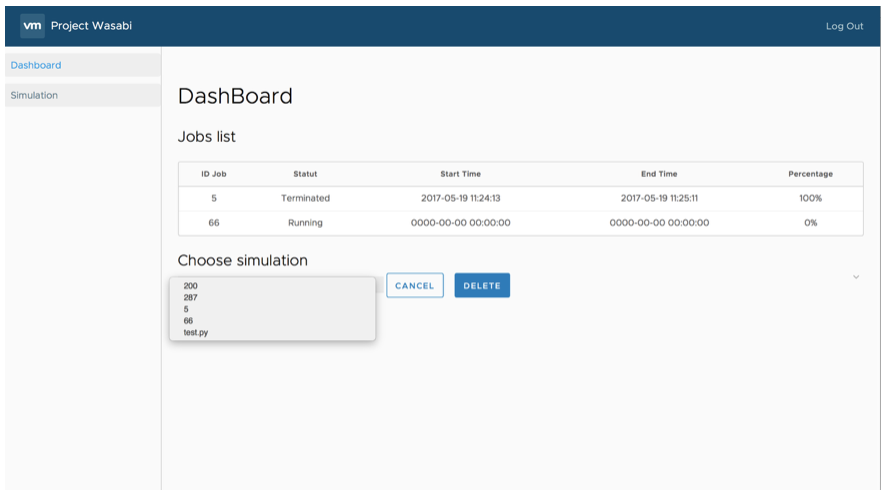
Figure 5: Screenshot of Job Management on the WebBoard
3. Information
3.1 The needs
- Language(s):
- C / Python for the client and servers
- Python pour l’application Wasabi
- Angular2, PHP, HTML, CSS pour l’interface
- Compiler(s): gcc
- Library(ies): none
- System(s): Linux
- Memory: Up to 2 GB.
- Disk: From a few MB to hundreds of GB
- Mean execution time: From a few seconds to several days.
- Databases : Yes